Dark matter of the genome, a safeguard of genetic integrity
Commentary on Fabrizio d'Adda di Fagagna's paper published on Nature
August 2013
With the advances of DNA sequencing technologies, the realization that 99% of the human genome is non-protein coding has highlighted the under-appreciated potential that lies within the dark matter of the genome. More strikingly, the recent ENCODE project estimated that the number of expressed non-coding genes, so called pervasive transcripts, is now more prominent than the number of coding genes [1]. Thus, understanding how non-coding transcription can be associated with particular function in vivo is an area that is subject of debate [2] and deserves deeper investigations. In particular it is important to explore how the pervasive transcription that generates thousands of non-coding transcripts can regulate gene expression and have broad impacts on development and disease in many organisms [3]. Long and small non-coding RNAs have no protein-coding potential and do not overlap with characterized classes of non-coding RNAs [4, 5]. In contrast to RNAi, miRNA and piRNA-mediated gene regulation, the novel classes of long and short ncRNA are still poorly defined whether it is for their putative regulatory role(s) or for their systematic catalog in living cells. In addition, if the majority of the ncRNA do not have function yet associated with their expression, gene regulation-mediated by ncRNA remains the most studied activity, leaving as terra incognita the role of the ncRNA on general DNA maintenance and chromosome stability.
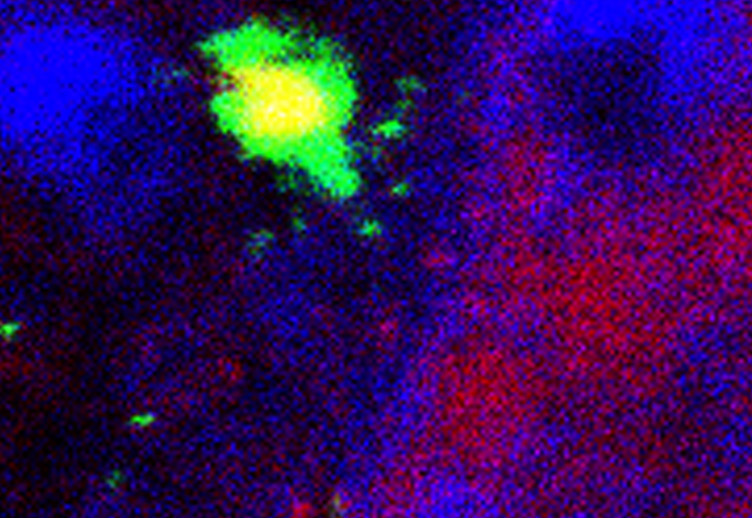
The team of Fabrizio d'Adda di Fagagna at IFOM, seeking for factors involved in DNA damage signaling pathways, explored why diverse human cells became sensitive to DNA damage when DICER and DROSHA, the two enzymes processing small ncRNA, are absent. Using high-throughput sequencing approaches in collaboration with the RIKEN institute in Japan, combined with appropriate genomic engineering on human and invertebrate systems, they identified the presence of novel small ncRNA, that they coined the DNA Damage RNA (DDRNA). These RNAs are produced from the DNA damage site [9] and necessary for signaling the damage to the DNA repair machinery. Indeed, impairing the biogenesis of the DDRNA results in cells enabled to repair and cell-cycle arrested. Strikingly, the very same damaged cells when grown in presence of a set of artificially produced DDRNA corresponding to the DNA damage site, recover their ability to repair their broken DNA. The authors propose that the DDRNA could trigger the molecular alarms through which the cell detects the problem and resolves it by repairing the damage. The exact molecular mechanism behind this DDRNA signaling pathway should be explored in the future.
Non-coding RNAs (ncRNAs) are involved in an increasingly recognized number of cellular events. Some ncRNAs are processed by DICER and DROSHA RNases to give rise to small double-stranded RNAs involved in RNA interference (RNAi). The DNA-damage response (DDR) is a signalling pathway that originates from a DNA lesion and arrests cell proliferation3. So far, DICER and DROSHA RNA products have not been reported to control DDR activation. Here we show, in human, mouse and zebrafish, that DICER and DROSHA, but not downstream elements of the RNAi pathway, are necessary to activate the DDR upon exogenous DNA damage and oncogene-induced genotoxic stress, as studied by DDR foci formation and by checkpoint assays. DDR foci are sensitive to RNase A treatment, and DICER- and DROSHA-dependent RNA products are required to restore DDR foci in RNase-A-treated cells. Through RNA deep sequencing and the study of DDR activation at a single inducible DNA double-strand break, we demonstrate that DDR foci formation requires site-specific DICER- and DROSHA-dependent small RNAs, named DDRNAs, which act in a MRE11–RAD50–NBS1–complex–dependent manner (MRE11 also known as MRE11A; NBS1 also known as NBN). DDRNAs, either chemically synthesized or in vitro generated by DICER cleavage, are sufficient to restore the DDR in RNase-A-treated cells, also in the absence of other cellular RNAs. Our results describe an unanticipated direct role of a novel class of ncRNAs in the control of DDR activation at sites of DNA damage. [PMID: 22722852]
Interestingly, two other teams, using two different model organisms, drosophila and Arabidopsis thaliana reached independently similar conclusions. Thus this novel DDRNA signaling pathway may have broad implications in various organisms. In drosophila, DDRNA are generated from the DNA double-strand breaks at the end of chromosome, resulting from an active sense/antisense transcription and leading to gene silencing at the vicinity of the damage [10]. In plants, DSB-induced small RNAs, similar to DDRNA, are potentiated by argonaute for an unknown process [11]. Although in both studies the direct roles of DDRNA have not been explored in details, these works suggest that DDRNA is a conserved and widespread mechanism that relays the DNA repair machinery, opening new avenues for the function of small ncRNA in DNA maintenance regulation.
The notion that the dark matter of the genome represents a tremendous resource for various functions is now coming into a reality. It would not be surprising to soon realize that beyond DNA processes even unsuspected cellular functions will be demonstrated to be modulated by ncRNA. (nc)RNA should thus take a full space as a major actor in the molecular biology of the cell and the genetic world [12, 13].
References
- [1] Djebali, S., et al. Nature, 2012. 489(7414): p. 101-8.
- [2] Clark, M.B., et al. PLoS Biol, 2011. 9(7): p. e1000625; discussion e1001102.
- [3] Tisseur, M., et al. Biochimie, 2011. 93(11): p. 1889-96.
- [4] Guttman, M., et al. Nature, 2009. 458(7235): p. 223-7.
- [5] Khalil, A.M., et al. Proc Natl Acad Sci U S A, 2009. 106(28): p. 11667-72.
- [6] Rinn, J.L., et al. Annu Rev Biochem, 2012. 81: p. 145-66.
- [7] Lee, H.C., et al. Nature, 2009. 459(7244): p. 274-7.
- [8] Zhang, Z., et al. Genes Dev, 2013. 27(2): p. 145-50.
- [9] Francia, S., et al. Nature, 2012. 488(7410): p. 231-5.
- [10] Michalik, K.M., et al. Nucleic Acids Res, 2012. 40(19): p. 9596-603.
- [11] Wei, W., et al. Cell, 2012. 149(1): p. 101-12.
- [12] Benner, S.A., et al. Science, 1991. 252(5010): p. 1232.
- [13] Waldrop, M.M. Science, 1989. 246(4935): p. 1248-9.