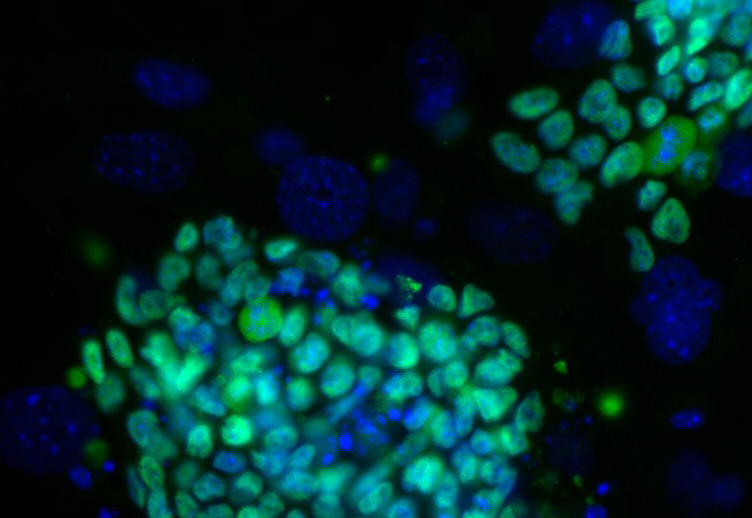
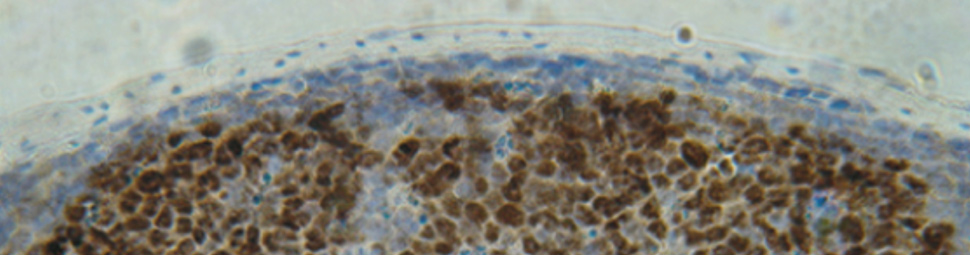
Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
Commentary on Stefano Casola's paper published on PLoS genetics
September 2014
The beginning of the 21st century will most likely be remembered in biomedical history as the time when stem cells took center stage, thanks to the incredible discovery of induced pluripotency. Immediately after the now seminal 2006 report of Yamanaka et al., scientists around the world started asking how a somatic cell could change its identity in such a dramatic fashion. It was suspected - in part thanks to the "rediscovered" Waddington's epigenetic landscape - that chromatin modifiers may be mechanistically involved in reprogramming. In recent years, several histone posttranslational modifications including methylation, acetylation, phosphorilation and others have all been implicated in gene regulation, and whether or not any of them participate in the process of induced pluripotency has become a topic of intense research.
Transcription factor (TF)-induced reprogramming of somatic cells into induced pluripotent stem cells (iPSC) is associated with genome-wide changes in chromatin modifications. Polycomb-mediated histone H3 lysine-27 trimethylation (H3K27me3) has been proposed as a defining mark that distinguishes the somatic from the iPSC epigenome. Here, we dissected the functional role of H3K27me3 in TF-induced reprogramming through the inactivation of the H3K27 methylase EZH2 at the onset of reprogramming. Our results demonstrate that surprisingly the establishment of functional iPSC proceeds despite global loss of H3K27me3. iPSC lacking EZH2 efficiently silenced the somatic transcriptome and differentiated into tissues derived from the three germ layers. Remarkably, the genome-wide analysis of H3K27me3 in Ezh2 mutant iPSC cells revealed the retention of this mark on a highly selected group of Polycomb targets enriched for developmental regulators controlling the expression of lineage specific genes. Erasure of H3K27me3 from these targets led to a striking impairment in TF-induced reprogramming. These results indicate that PRC2-mediated H3K27 trimethylation is required on a highly selective core of Polycomb targets whose repression enables TF-dependent cell reprogramming.
[PMID 23468641]
In this regard the manuscript by Stefano Casola and Giuseppe Testa's labs represents a key advance in our understanding of how chromatin remodeling regulates reprogramming. Before their studies came to light it was generally accepted that a wave of dimethylation of lysine-4 of histone H3 (H3K4me2) was established at enhancers and promoters of genes important for pluripotency preceding reprogramming, but that chromatin state differed in established induced pluripotent stem cells (iPSC) which showed Polycomb-mediated trimethylation of lysine 27 of histone 3 (H3K27me3) (Boyer et al, 2006; Maherali et al, 2007; Mansour et al, 2012; Onder et al, 2012). Hence, a major role was attributed to the gene silencing activity of the PRC2 complex (mainly mediated by Ezh2) in establishing this H3K27me3 signature, which became the chromatin hallmark of iPSC. The elegant studies by the Casola and Testa's labs utilizing conditional Ezh2 knock-out cells, revealed that in actuality, Ezh2 and global H3K27me3 were dispensable for reprogramming to occur, except for a highly specific H3K27me3 mark on a defined core of Polycomb targets that ultimately enables nuclear reprogramming. More surprisingly, they showed for the first time that this gene repression mark was achieved through an alternative PRC2, most likely Ezh1, providing the first functional validation of the role of PRC2 in the establishment of pluripotency. This was further confirmed by their findings that downregulation of EED, an essential component of PRC2 significantly reduced H3K27me3 and prevented reprogramming.
The concepts illuminated by Stefano and Giuseppe's manuscript provide the basis for and in some cases explained several recent articles (Gafni et al, 2013; Malouf et al, 2013; Tiwari et al, 2013; Ding et al, 2014), confirming the significance and importance of their original findings. Indeed, the fact that H3K27me3 marks are retained in specific hotspots and that is mediated independently of Ezh2, opens new exciting opportunities to advance our understanding of the basic processes of reprogramming and provides novel targets for the manipulation of cell fate. I have been lucky to know Stefano and Giuseppe for many years and I am proud to have played a small part in this seminal paper.