TALE homeodomain transcription factors: super-regulators of the genome transcriptional specificity
Commentary on Francesco Blasi's paper published on Cell reports
July 2014
Transcription factors (TF) are proteins that regulate gene expression through recognizing and binding specific DNA sequences on their target genes. Two major classes of TFs can be distinguished; general TFs, which are common to a majority of genes, and specific TFs, which specifically regulate certain genes in space and time during embryonic development and adult life. Homeodomain TFs belong to this last family and are characterized by the presence of a DNA binding motif highly conserved in evolution termed homeodomain. Nowadays it is estimated that there are more than 500 homeodomain protein-encoding genes in vertebrates. Homeodomain proteins play essential roles in specifying body parts and in determining cell identities, differentiation and proliferation, during embryonic development and adult tissue homeostasis.
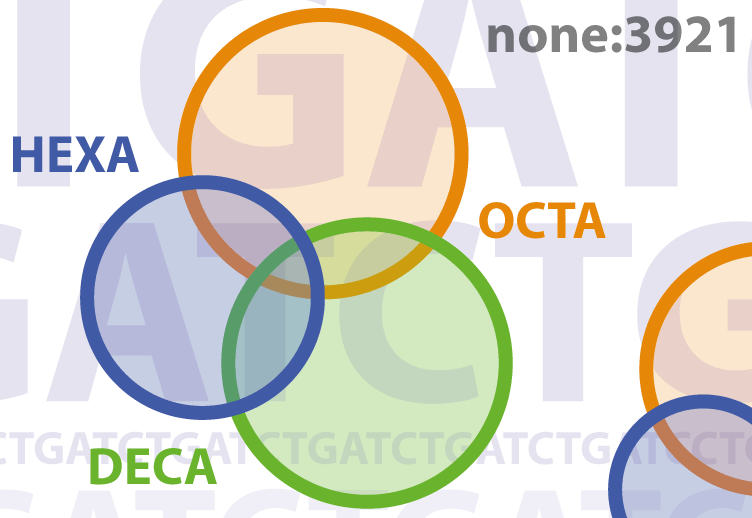
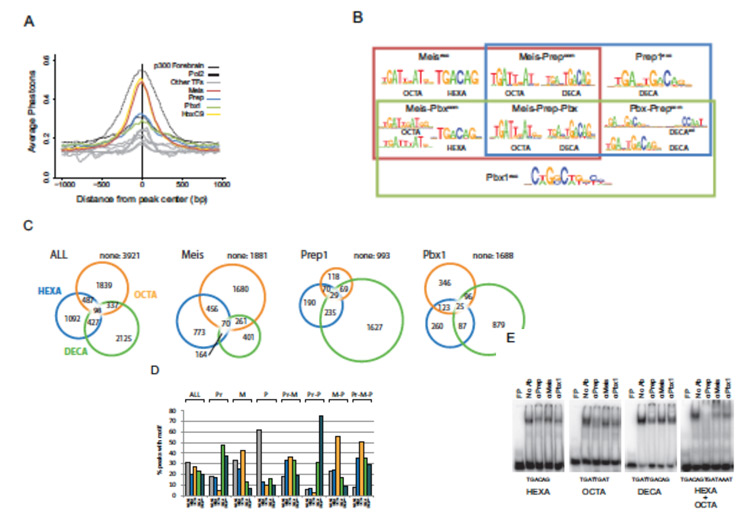
The interactions of Meis, Prep, and Pbx1 TALE homeoproteins with Hox proteins are essential for development and disease. Although Meis and Prep behave similarly in vitro, their in vivo activities remain largely unexplored. We show that Prep and Meis interact with largely independent sets of genomic sites and select different DNA-binding sequences, Prep associating mostly with promoters and housekeeping genes and Meis with promoter-remote regions and developmental genes. Hox target sequences associate strongly with Meis but not with Prep binding sites, while Pbx1 cooperates with both Prep and Meis. Accordingly, Meis1 shows strong genetic interaction with Pbx1 but not with Prep1. Meis1 and Prep1 nonetheless coregulate a subset of genes, predominantly through opposing effects. Notably, the TALE homeoprotein binding profile subdivides Hox clusters into two domains differentially regulated by Meis1 and Prep1. During evolution, Meis and Prep thus specialized their interactions but maintained significant regulatory coordination.
[PMID 23602564]
The homeodomain was named after its discovery in genes belonging to the Drosophila bithorax and antenapedia gene complexes, whose mutations lead to so-called homeotic transformations, which transform a body part into the appearance of another. The genes within these complexes, named Hox, confer segmental identity along the main antero-posterior (A-P) embryonic axis. Hox gene complexes are conserved in evolution at the molecular and functional levels. The ability of Hox proteins to bind their target DNA depends critically on their interaction with cofactors of the TALE (Three Aminoacids Length Extension) homeodomain subfamily. There are two TALE subfamilies playing this role in vertebrates, the Pbx subfamily with 4 genes and the Meis/Prep with 3 Meis and and 2 Prep genes (discovered by Francesco Blasi in 1988). Further analyses led to the identification of additional roles for Pbx and Meis/Prep TFs as cofactors of several other families of tissue-specific TFs. Not surprisingly, genetic alterations in members of the TALE-Hox network correlate both with congenital and acquired diseases such as cancer. Given their ability to regulate the activity of sets of TFs that control specific pathways, the TALE TFs occupy a very high position in the genome regulatory hierarchy and can be regarded as super-regulators of the genome transcriptional specificity.
Most of the knowledge accumulated on the role of Meis-Prep and Pbx biochemical interactions had been addressed in vitro and was derived form the analysis of a few specific cases of gene regulation. The picture emerging for these previous studies mostly suggested similar DNA binding specificity and redundant roles in interactions with Hox proteins/genes for Meis and Prep factors; however, this view was in conflict with the mutant phenotypes found respectively by Blasi in Prep1 mutants and by my own laboratory in Meis1 mutants. The situation required at this point a comprehensive and unbiased study of the in vivo specificity of TALE factors, something that could only be achieved through the coordination of several laboratories interested in this area. It was the initiative of Francesco Blasi that made this possible; we had recently obtained a COST grant for the coordination of research efforts across Europe towards the understanding of TALE homeodomain protein function, when I received a phone call from Francesco proposing a coordinated effort to achieve this important goal. He did not need much insistence to convince me of embarking on this project and we rapidly put together efforts from laboratories in Italy, Spain, Germany and Rusia that had in hands the reagents and expertise required for this ambitious project. Main actors of this effort were Dmitry Penkov from the University of Moscow, a former Blasi lab postdoc and specialist in protein-DNA interactions, and Daniel Mateos San Martín, a PhD student in my lab with a special talent for bioinformatics.
Together we could generate ChIP-seq data addressing the DNA binding landscape of a majority of Meis-Prep and Pbx TFs. Importantly this was achieved in vivo and targeting the endogenous native proteins. The studies were complemented with phenotypic and transcriptional analyses of several single-factor mutant and compound mutant mice. The informatics analysis of the huge amount of data generated indicated a very different picture from previous conceptions in the field; Meis and Prep factors, not only were not redundant but they showed complementary DNA-binding patterns, different DNA sequence preference and, most importantly, different cofactor-binding preferences. Not surprisingly, Meis/Prep compound mutants displayed no cooperative interactions in any organ/tissue studied. The differences identified affected especially the ability of Meis/Prep to interact with Hox proteins; Meis was the preferred DNA binding partner for Hox proteins and, surprisingly, the nearly exclusive factor binding on Hox clusters for Hox gene regulation. The structural analysis of the binding profiles was not less surprising; while Prep was revealed as promoter-binding transcription factor, Meis bound with a strong preference to promoter-remote regions. Finally, the type of genes bound by each factor revealed that Prep bound mostly genes acting on basic cellular functions, while Meis preferentially bound developmentally regulated genes.
A recent example of the application of the knowledge generated in this work derives from the observation that a significant set of genes co-regulated by Meis and Prep displayed antagonistic regulation by these factors. Based on these observations the group of Francesco Blasi has recently described in PNAS the antagonistic activity of Meis1 and Prep1 in tumor formation. This study has thus modified the way we understand the regulatory interactions within a family of TFs essential for animal development and adult tissue homeostasis. Thanks to the vision of Francesco Blasi and a fruitful collaborative effort, the knowledge generated in this work is paving the way to new discoveries and will likely have a long-standing impact in several fields of research.