Seeing is believing: catching template-switching in the act
Commentary on Dana Branzei's paper published on Nature Structural & Molecular Biology
March 2015
Every time a cell replicates its genome, it runs the risk of breaking it. Replication involves copying millions to billions of nucleotides, and this must be done faithfully if two identical versions of the genome are to be passed on to the next generation. Evolution has selected for DNA replication enzymes that accomplish this task with efficiency and fidelity, but on occasion even this marvelous machine breaks down. DNA damage, both spontaneous and induced, creates lesions that cannot be copied, resulting in breaks in the vicinity of the replication fork. If these breaks cannot be repaired rapidly and faithfully, they can lead to mutations and rearrangements that alter genome content and place critical genes in inappropriate regulatory contexts. These are a leading source of genetic disease and a leading cause of cancer.
Template switching (TS) mediates damage bypass via a recombination-related mechanism involving PCNA polyubiquitination and polymerase δ–dependent DNA synthesis. Using two-dimensional gel electrophoresis and EM, here we characterize TS intermediates arising in Saccharomyces cerevisiae at a defined chromosome locus, identifying five major families of intermediates. Single-stranded DNA gaps of 150–200 nt, and not DNA ends, initiate TS by strand invasion. This causes reannealing of the parental strands and exposure of the nondamaged newly synthesized chromatid, which serves as a replication template for the other blocked nascent strand. Structures resembling double Holliday junctions, postulated to be central double-strand break–repair intermediates but so far visualized only in meiosis, mediate late stages of TS before being processed to hemicatenanes. Our results reveal the DNA transitions accounting for recombination-mediated DNA-damage tolerance in mitotic cells and replication under conditions of genotoxic stress. [PMID 25195051]
As a consequence, mechanisms have evolved that preserve genome integrity by tolerating replication-blocking DNA lesions. Two are generally used: the bypass of lesions by error-prone polymerases that introduce mutations at the site of damage; and error-free mechanisms that use homologous recombination to copy correct information from an undamaged template, creating a patch that bypasses the lesion and restores genome integrity and function at the site of damage. This latter mechanism, called template switching, is a major form of lesion bypass in normal cells, as evidenced by findings that most DNA damage is repaired without mutation or genome alteration.
Mechanisms of template switching have been the subject of intense focus, in particular at the IFOM. Much progress has been made in understanding the proteins and enzymatic activities involved, using genetic and cytological approaches, through in vitro biochemical studies, and using gel electrophoresis-based approaches that allow inference about template switching intermediate structures. However, until now, the somewhat indirect nature of these approaches has produced incomplete understanding. In particular, progress has been limited by an inability to distinguish between two alternative models for template switching. In one, a stalled replication fork regresses to form a four-armed DNA structure in which DNA synthesis, followed by fork restoration, creates a patch to cover a lesion. In the second, the fork does not stall, but leaves behind single-strand DNA gaps, which are used by homologous recombination activities to prime new DNA synthesis, generating a lesion-covering patch. Both models can accommodate existing genetic and biochemical data, and both are expected to produce the intermediate structures detected on gels. Because template switching occurs at relatively low frequencies, and at many places in the genome, distinguishing between these and other models has been stymied by the fact that we have not been able to actually see the DNA molecules that are engaged in template switching—until now.
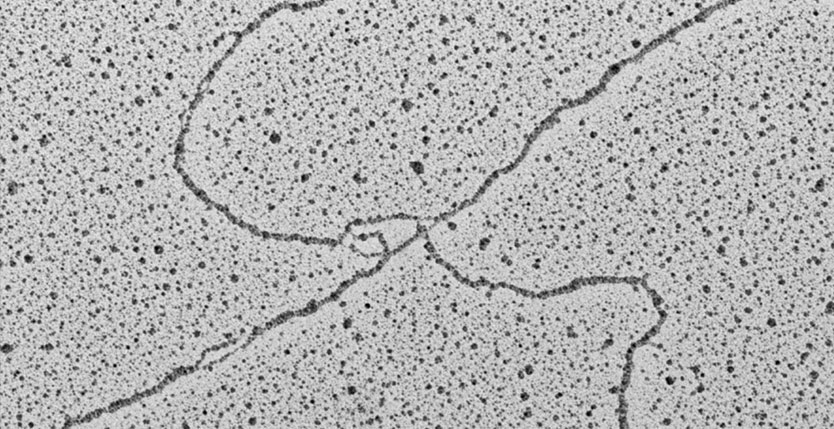
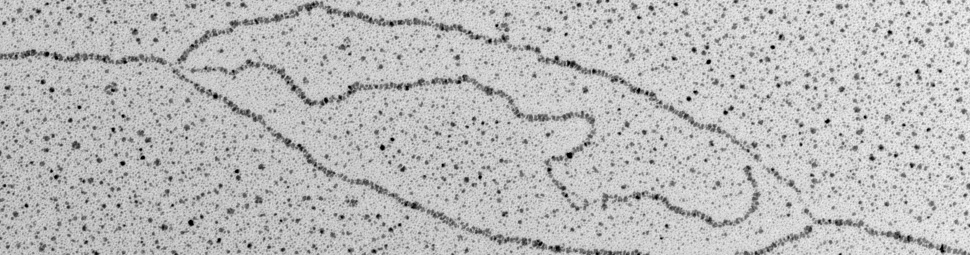
The work that filled this gap was performed at the IFOM by the group of Dana Branzei and her collaborators. They solved the problems of low frequency and multiple locations by creating a budding yeast strain that contains a high-copy minichromosome, present in dozens of copies in a single cell, that can be directly seen on agarose gels. They then used methy methanesulfonate to induce template switching at high levels, and combined methods to stabilize template-switching intermediates and methods to separate these intermediates from the bulk of genomic DNA on 2-dimensional agarose gels. Branzei and colleagues were then able to extract these intermediates from gels and directly visualize their structures in the electron microscope.
This remarkable study represents the first time in that eukaryotic repair intermediates have been visualized on a specified DNA molecule, and it has provided unprecedented insight into template switching mechanisms. In particular, the spectrum of intermediate structures Branzei and colleagues detect is inconsistent with fork regression being the dominant template switching mechanism. Rather, they point to mechanisms where DNA at a single-strand gap, left behind by the replication fork, interacts with homologous DNA on the undamaged sister chromatid and initiates new synthesis, ultimately liberating an intact strand that repairs the damaged chromosome. One prediction of this model is that repair products frequently should be linked by two intertwined single strands in a structure called a hemicatenane. Previous work at the IFOM has shown that these structures can frequently be produced when template switching is induced.
In summary, Branzei and colleagues have provided insight into template switching at an unparalleled level of molecular detail. Their study points the way for future approaches, not just in the study of template switching, but also of other homology-based recombination processes that play additional important roles in preserving genome integrity.