FANCM – a new cancer susceptibility gene?
Commentary on Paolo Peterlongo's paper published in Human Molecular Genetics.
August 2016
Breast cancer susceptibility genes
Over twenty years ago, the important breast cancer susceptibility genes, BRCA1 and BRCA2 were discovered. Since this time, the search for “BRCA3” has been vigorously pursued by many laboratories around the world. Several candidate susceptibility genes have been identified, but none match the unique properties held by mutations in BRCA1 and BRCA2 – that is, being both highly penetrant and quite frequent in most populations. Nevertheless, an international collaborative group, including Dr. Paolo Peterlongo and myself, showed in 2014 that PALB2 is a bona fide cancer susceptibility gene associated with a clinically significant risk for breast (and possibly other) cancers.
The challenge now is to validate the many other candidate breast cancer susceptibility genes that have emerged from large-scale studies in Europe and North America. One such study has been led by Dr. Paolo Peterlongo, from IFOM, the FIRC Institute of Molecular Oncology, in Milan and has uncovered an association between the presence of likely pathogenic mutations in the gene encoding FANCM, a member of the Fanconi Anemia (FA) protein family, and risk for breast cancer.
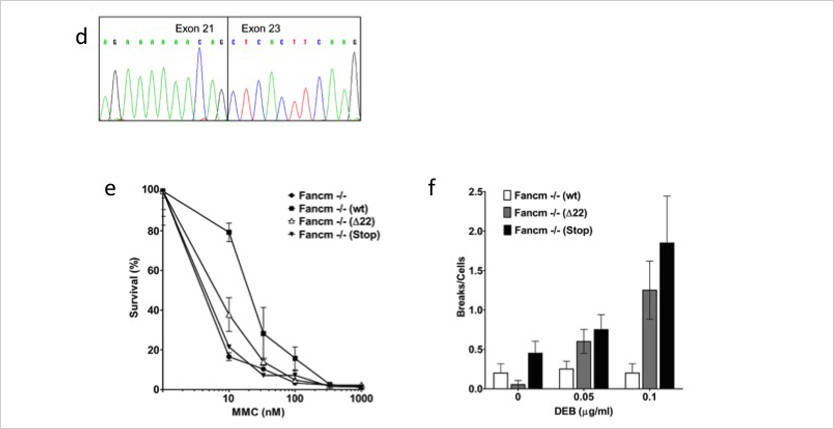
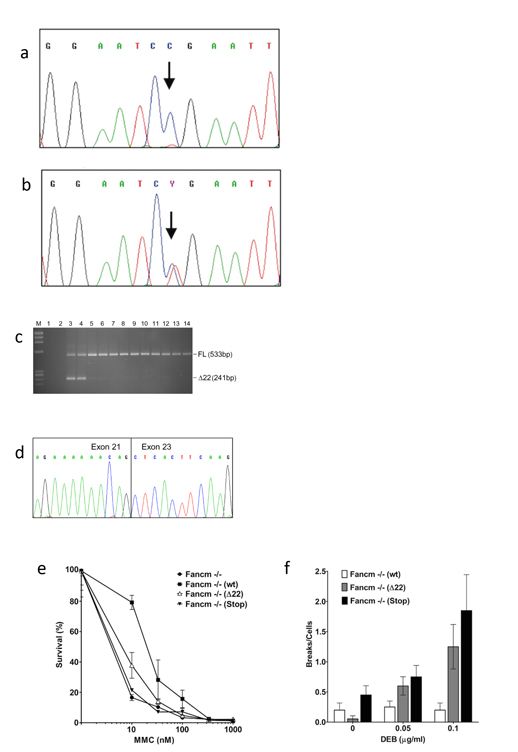
Numerous genetic factors that influence breast cancer risk are known. However, approximately two-thirds of the overall familial risk remain unexplained. To determine whether some of the missing heritability is due to rare variants conferring high to moderate risk, we tested for an association between the c.5791C>T nonsense mutation (p.Arg1931*; rs144567652) in exon 22 of FANCM gene and breast cancer. An analysis of genotyping data from 8635 familial breast cancer cases and 6625 controls from different countries yielded an association between the c.5791C>T mutation and breast cancer risk [odds ratio (OR) = 3.93 (95% confidence interval (CI) = 1.28-12.11; P = 0.017)]. Moreover, we performed two meta-analyses of studies from countries with carriers in both cases and controls and of all available data. These analyses showed breast cancer associations with OR = 3.67 (95% CI = 1.04-12.87; P = 0.043) and OR = 3.33 (95% CI = 1.09-13.62; P = 0.032), respectively. Based on information theory-based prediction, we established that the mutation caused an out-of-frame deletion of exon 22, due to the creation of a binding site for the pre-mRNA processing protein hnRNP A1. Furthermore, genetic complementation analyses showed that the mutation influenced the DNA repair activity of the FANCM protein. In summary, we provide evidence for the first time showing that the common p.Arg1931* loss-of-function variant in FANCM is a risk factor for familial breast cancer.
[PMID 26130695]
How was FANCM discovered?
FANCM, initially known as FAAP250, was identified in 2005 as a human ortholog of the ancient bacterial DNA repair protein, Hef1, via mass spectrometry analysis of FA protein complexes. While its role as a true FA protein is questionable (see below) it does have key DNA repair functions, briefly outlined below. Since nearly all existing breast cancer susceptibility genes appear to have roles in DNA repair, FANCM is a priori a candidate breast cancer susceptibility gene.
What does it do?
There are 19 FA associated proteins. Among many functions, it is believed that FANCM acts as a DNA translocase, and interacts with a number of partners to recognise stalled replication forks and activate the FA pathway. Other FA core components are then recruited to the DNA lesion. However, it is not clear that FANCM is a bona fide FA gene, since several homozygous mutation carriers have been found not to have any molecular or clinical signs of FA, and the only known carrier of biallelic mutations in FANCM was later found to have mutations in FANCA, another FA gene, as well.
How was FANCM linked to breast cancer?
A study of multiple-case breast cancer families identified a single affected woman who was heterozygous for a variant in FANCM known as c.5791C>T (rs144567652). Further analysis of several thousand cases suggested that this variant was associated with breast cancer risk, but the sample size, while large in the first study, was not definitive, because the variant was only seen in about 3 in 1000 women with breast cancer. A subsequent study, lead by Dr. Peterlongo, which analysed 8635 familial breast cancer cases and 6625 controls from several different countries for this single mutations c.5791C>T, found an association between this mutation and breast cancer risk [odds ratio = 3.9 (95% confidence interval = 1.3–12.1; P = 0.017)]. Functionally, it was shown that this mutation causes an out-of-frame deletion of exon 22, resulting from the creation of a de novo binding site for the pre-mRNA processing protein hnRNP A1; moreover, genetic complementation analyses showed that the c.5791C>T mutation can influence the DNA repair activity of FANCM. Along with another study, suggesting that a Finland-specific FANCM mutation is associated with triple negative breast cancer, these observations are quite compelling. Additional support for these data comes from a large study of 4034 cases of a total of 12 tumor types, published in Nature Communications in 2015, which revealed that using a total frequency test FANCM was significantly enriched for germline truncating mutations in breast cancer – in fact, FANCM was the third most-frequently mutated gene, after BRCA1 and BRCA2, but this excess did not cross the 5% false discovery rate threshold.
Are FANCM mutations found in other cancers/conditions?
In the abovementioned Nature Communications study, germline FANCM mutations were unexpectedly frequent in persons with head and neck squamous cell carcinoma (HNSCC) and clear cell carcinoma of the kidney (RCC). Many of the germline FANCM mutations were associated with LOH, suggesting that they might be biologically important. Notably, using a Wilcoxon rank-sum test, FANCM mutations were associated with a greater number of somatic mutations in HNSCC and RCC, than in tumors without FANCM mutations.
Where do we go next?
Validation of FANCM as a breast cancer susceptibility gene will require much larger studies than already been conducted; to disprove that BRIP1 was a breast cancer susceptibility gene required genotyping for one variant in a combined total of 91,000 cases and controls, and full sequencing of the gene in a further 24,000 women. These types of sample sizes are needed for rare variants with modest effects. Thus, along with functional studies, further sequencing is in order, but Dr. Peterlongo has certainly identified a strong candidate breast cancer susceptibility gene – no mean feat.