A key insight on the enzymatic activity of AID
A commentary to Svend Petersen-Mahrt's paper published on Journal of Experimental Medicine
June 2013
DNA alterations that lead to cancer are often generated via chemical or physical damage to our genomes and can be caused, for example, by smoking or UV light, which induce defined lesions into DNA. At the heart of our own defences against cancer lie a multitude of biochemical pathways that have evolved to repair this damage, keeping our genomes healthy. However, to ward off the invasive pathogens such as bacteria and viruses, evolution has also designed a system in which intentionally-induced DNA damage is a key step in producing a functional immune system.
Antibody diversification requires the DNA deaminase AID to induce DNA instability at immunoglobulin (Ig) loci upon B cell stimulation. For efficient cytosine deamination, AID requires single-stranded DNA and needs to gain access to Ig loci, with RNA pol II transcription possibly providing both aspects. To understand these mechanisms, we isolated and characterized endogenous AID-containing protein complexes from the chromatin of diversifying B cells. The majority of proteins associated with AID belonged to RNA polymerase II elongation and chromatin modification complexes. Besides the two core polymerase subunits, members of the PAF complex, SUPT5H, SUPT6H, and FACT complex associated with AID. We show that AID associates with RNA polymerase-associated factor 1 (PAF1) through its N-terminal domain, that depletion of PAF complex members inhibits AID-induced immune diversification, and that the PAF complex can serve as a binding platform for AID on chromatin. A model is emerging of how RNA polymerase II elongation and pausing induce and resolve AID lesions.. [PMID: 23008333]
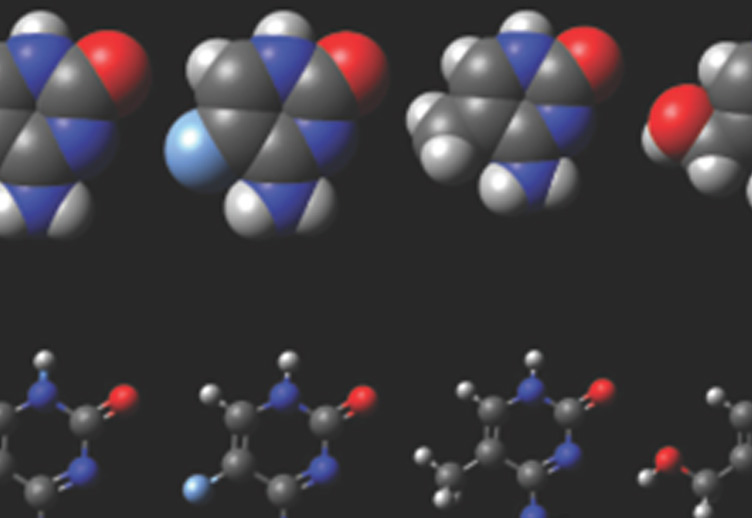
The work from Dr. Petersen-Mahrt's laboratory provides a key insight into how this intentional DNA damage is introduced into the antibody genes, which in turn could also provide insight how such damage could lead to cancer. The breakthrough came from discovering how a small protein, AID, functions within the nucleus. AID is a member of a family of proteins that induce DNA damage by removing an amino group (deaminating) from the base cytosine in our DNA. If this deamination is not corrected, then the deaminated cytosine will miscode for another base during DNA copying and mutations will arise. Should AID lose its focus on its usual target (the antibody genes), then deaminated cytosines will be generated throughout the genome and cause mutations predisposing to cancer development. Using a novel approach to isolating AID protein from immune cells, Dr. Petersen-Mahrt's group was able, for the first time, to demonstrate the existence of a particular protein complex on chromatin containing AID and other members of the gene transcription machinery. By identifying an essential protein (PAF1) of the transcription machinery as an associated partner of AID, Dr. Petersen-Mahrt and his team were able to provide a model as to how the cell controls AID activity. During the act of transcribing a gene, the RNA polymerase will slow down, stall and restart - processes influenced by PAF1. If AID associates with RNA polymerase during this time, the deaminated cytosines might not be repaired but could give rise to mutations and recombination. Such a mechanism is consistent with molecular observations made by other laboratories in the field. A group from Rockefeller University had uncovered a complex that functions upstream of PAF1, while work of groups from Kyoto University and Harvard University identified downstream connections. Dr. Petersen-Mahrt was able to confirm all the proposed connections, which should greatly assist the emergence of a coherent view of how AID is retained and functions at the antibody gene loci.
The role of AID in causing cancer has been well documented, but suitable pharmacological targets, aside from AID, have not been identified: PAF1 may prove a possible target. Because AID has been implicated in the development of chemo-drug resistance in patients, interrupting the PAF1-AID association might be a means to control AID mutability during therapy. The on-going work in Dr. Petersen-Mahrt is geared towards using high-throughput analysis to identify AID and AID function inhibitors.
Evolutionary offsprings of AID (APOBEC3 family of proteins) are known to inhibit viral infections such as HIV. A recent report has shown that PAF1 has an important function in HIV restriction, yet a functional mechanism was not proposed. In light of Dr. Petersen-Mahrt lab's finding, it could be that the interaction of the AID/APOBEC family with the RNA transcription machinery is ancient and conserved, and pharmacological characterisation could be beneficial for cancer as well as HIV therapy.